Shaking the Rust Off Python Redox
This post is an update to Shaking the Rust Off Python
I would like to first thank Dr. Michael Lazear and Dr. Rob Patro for their wonderful feedback on my recent blog post. Dr. Lazear has generously shown that there is a better implementation that can deliver a blazingly fast compute time by using “map-reduce” with worker pool in the rayon package of Rust. In swift response to Dr. Lazear’s suggestion, Dr. Patro initially confirmed that the suggested implementation delivers a substantial improvement over the Python baseline (also thank you for teaching me about the ‘—release’ parameter for ‘maturin develop’). This approach indeed leads to a 10x improvement in Rust+Python over the pure Python implementation.
1. Updated Implementation (Rust+Python)
Here is the updated Rust+Python (multithreaded) implementation that uses map-reduce in the rayon package (the credit goes to Dr. Lazear for the original source code):
lib.rs#[pyfunction] fn count_kmers_multithread_fx_hashmap_improved(sequences: Vec<String>, k: usize, num_threads: usize) -> Py<PyAny> { let pool = rayon::ThreadPoolBuilder::new() .num_threads(num_threads) .build() .unwrap(); let hm = pool.install(|| { let hm = sequences .par_iter() .map(|sequence| { let end = sequence.len() - k + 1; let mut map: FxHashMap<_, i32> = FxHashMap::default(); for i in 0..end { *map.entry(&sequence[i..i+k]).or_insert(0) += 1; } map }) .reduce( || FxHashMap::default(), |mut acc, x| { for (k, v) in x { *acc.entry(k).or_insert(0) += v; } acc }, ); return hm; }); Python::with_gil(|py| hm.to_object(py)) } #[pymodule] fn scripts(_py: Python, m: &PyModule) -> PyResult<()> { m.add_function(wrap_pyfunction!(count_kmers_multithread_fx_hashmap_improved, m)?)?; Ok(()) }
count_kmers_rust_multithread_fx_hashmap_improved.pyimport pysam import time import pandas as pd import scripts def count_kmers_rust_multithread_fx_hashmap_improved( fasta_file: str, num_processes: int, sequence_chunk: int, k: int, chromosomes: list) -> pd.DataFrame: """ Counts all possible k-mers in a FASTA file. Parameters ---------- fasta_file : FASTA file. num_processes : Number of processes to multiprocess FASTA file. sequence_chunk : Sequence chunk. k : k-mer k. chromosomes : Chromosomes to count. Returns ------- df_kmers : DataFrame of k-mers with the following columns: 'k_mer', 'count' duration_1 : Duration of step 1. duration_2 : Duration of step 2. duration_3 : Duration of step 3. duration_total : Total duration. """ # Step 1. Create tasks start_time_1 = time.time() fasta = pysam.FastaFile(fasta_file) sequences = [] for curr_chromosome in chromosomes: curr_chromosome_len = fasta.get_reference_length(curr_chromosome) for i in range(0, curr_chromosome_len, sequence_chunk): curr_start = i curr_end = curr_start + sequence_chunk if curr_end > curr_chromosome_len: curr_end = curr_chromosome_len curr_chromosome_seq = fasta.fetch(reference=curr_chromosome, start=curr_start, end=curr_end) sequences.append(curr_chromosome_seq.upper()) end_time_1 = time.time() # Step 2. Run start_time_2 = time.time() results = scripts.count_kmers_multithread_fx_hashmap_improved(sequences, k, num_processes) end_time_2 = time.time() # Step 3. Merge dictionaries into one start_time_3 = time.time() df_kmers = pd.DataFrame(results.items()) df_kmers.columns = ['k_mer', 'count'] df_kmers.sort_values(['count'], ascending=False, inplace=True) end_time_3 = time.time() return df_kmers, \ end_time_1 - start_time_1, \ end_time_2 - start_time_2, \ end_time_3 - start_time_3, \ end_time_3 - start_time_1
2. Updated Benchmark Test Results
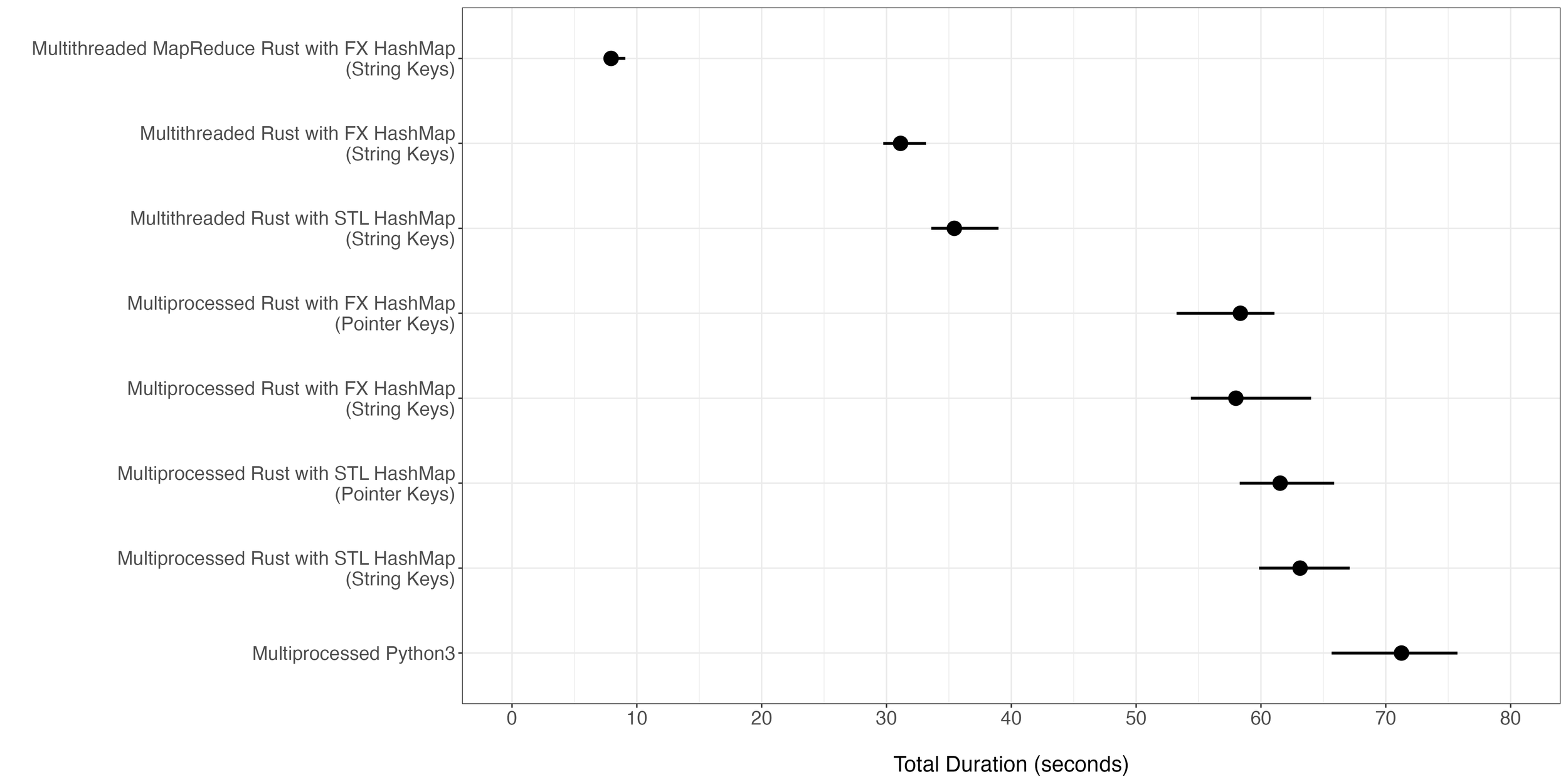
The benchmark test configuration was kept the same as the original post (9-mer counting of chromosome 1 in hg38 with 24 cores with sequences of 1 million nucleotides in length). The Rust implementation using map-reduce led to a 9.6x improvement from the pure Python equivalent (Figures 1 and 2). This was also a substantial improvement compared to the former Rust implementation that relied on a multithreading pool. Note that the benchmark tests were run again on a different node with less powerful cores, which is why you see a different range of durations compared to the original post. Nevertheless, the comparative speed improvement from Python to Rust is conserved from the previous test to the current test.
Figure 1. Total execution time of different implementations for the task of 9-mer counting for hg38 chromosome 1. For each implementation, 20 iterations were run with each iteration using 24 cores. The dot in the middle of each bar represents the median total duration while the ends of the bar represent the minimum and the maximum duration time observed.
Figure 2. Average execution times of each of the three steps observed in the same tests for Figure 1.
3. Conclusions
I have learned a great deal from receiving feedback from the much active scientific/bioinformatics community on Twitter at large. So thank you, Dr. Lazear and Dr. Patro, for your help and time. I would like to also thank my co-advisor and principal investigator, Dr. Alex Rubinsteyn, for sharing the post on Twitter in the first place. You were right, Chief!
The experience of writing a blog post about a topic that I am just beginning to learn and then tweeting it has inadvertently become a manifestation of Cunningham’s Law. I suspect that Alex knew about it all along. In any case, I have learned a great deal about using Rust and human behavior. I promise to use these newly acquired skillsets sparingly and responsibly.