Tricking Nextflow's Caching System (to Drastically Reduce Storage Usage)
Introduction
Policing storage usage is an unfortunate, but necessary aspect of bioinformatics. This is due to both the relatively large files workflows generate and the sheer number of files associated with a potentially large set of samples. It is easy for this usage to get out of hand in cases where strict controls are not in place. I am, unfortunately, guilty as charged when it comes to the sin of excess storage usage and I attribute most of this usage to Nextflow's picky caching system.
This problem is not specific to me and my colleagues. A Nextflow GitHub issues thread regarding this topic recently celebrated its fifth birthday and Ben Sherman (@bentsherman) has, as of the writing of this post, started laying the ground work to allow Nextflow to utilize temporary intermediate files.
Our lab has been tackling storage issues since we started using Nextflow DSL2 way back in November 2019. We were unable to "trick" Nextflow into caching near-zero-sized files despite our best efforts. Luckily, Stephen Ficklin and colleagues, authors of GEMmaker, have developed a clever solution to this problem.
This blog post is to show examples explaining the utility of this approach, provide a syntactical tutorial showing implementation examples, and describe the limitations and pitfalls I encountered while implementing it. Before we discuss the technical aspects of intermediate file cleaning in Nextflow workflows, let's further elaborate on the need for this functionality.
Why?
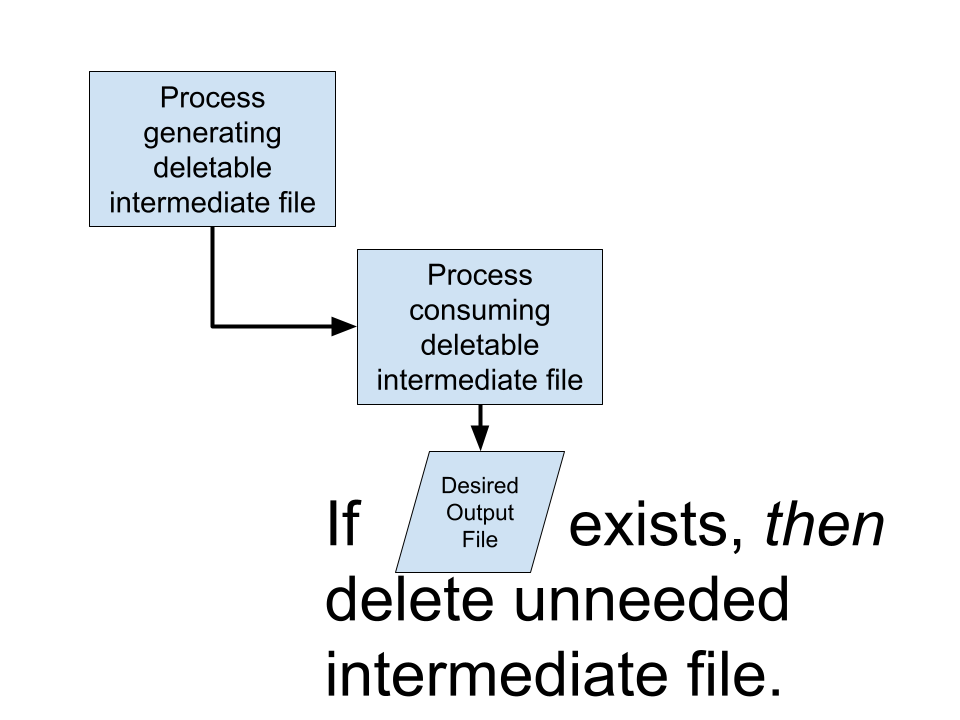
Consider the following workflow and its associated files:
In our case, we are only required to retain the first (raw FASTQs) and last (fully processed BAMs) files for the purposes of reproducibility and to satisfy downstream workflow inputs, respectively. Nevertheless, Nextflow in its current form requires the four intermediate files to be available on the filesystem for workflow caching purposes.
Now consider the following workflow and its associated files:
Deleting intermediate results in a 75% reduction in storage requirements for the same eventual workflow outcome. This allows for more samples to be processed using the same storage with less downtime resulting from storage maintenance. This optimization can be taken to the extreme by having the fully processed BAM (the star shaped element in our diagram) deleted once your workflow generates its final outputs (e.g. transcript counts, VCFs, etc.). There are, of course, trade-offs to this strategy which we'll discuss in the Limitations and Pitfalls section.
How?
In a nutshell, the GEMmaker strategy works by creating a very small file that resembles the original file closely enough that Nextflow cannot tell the difference. The bash script which performs this action is available in the GEMmaker Github repositorty. Briefly, the script stats the file, records the relevant statistics, creates a sparse file and modifies the sparse file's logical size, modification time, and access time to reflect those of the original file. The result is an indistinguishable (by Nextflow, anyways) file to serve as a placeholder for your previously large intermediate file.
By This file was made by User:Sven
CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=4067353
When?
Hopefully one can appreciate the value of the sparse file strategy when applied to running large scale Nextflow workflows. One may be tempted to use this hammer to all of the "nails" in their workflows. Unfortunately, every rose has its thorn. Specifically, the timing of intermediate file cleaning is relatively delicate. Any downstream processes or workflows that utilize the cleanable intermediate files must be completed prior to cleaning or your workflow will fail due to the prematurely deleted intermediate file. User will then be stuck re-running a portion of your workflow after debugging the cause.
The next aspect of "when?" is more philosophical -- is it better to delete as soon as possible or at the end of the workflow when all of your endpoint files are available? Should deletion occur within your top-level workflow (as in GEMmaker) or sub-workflows? This is a topic I've wrestled with while implementing intermediate file deletion in our neoantigen workflow LENS so I have thoughts on it that I can share in a future blog post assuming there's interest.
A Syntax Example
The GEMmaker implementation has been suggested as a potential workaround to commenters in Nextflow's GitHub Issue #452. I attempted to implement this strategy after strong (but understandable) insistence by our sysadmins that I use less storage. I initially found the logic a bit hard to follow (the code is clear, but I am a mere geneticist, not a software engineer!), so I wanted to create a verbose yet minimal example to explain the core logic. The code (trick_nextflow_cache.nf, clean_work_dirs.sh, and utilities.nf) described below can be found on GitHub: https://github.com/pirl-unc/tricking_nextflow_cache.
First, we'll demonstrate the functionality and then walk through the script.
We begin with an empty work directory:
[spvensko@c6145-2-9 tricking_nextflow_cache_example]$ du -hscL work/ 4.0K work 4.0K total
Next, we run our Nextflow script:
[spvensko@c6145-2-9 tricking_nextflow_cache_example]$ nextflow trick_nextflow_cache.nf -resume N E X T F L O W ~ version 21.10.6 Launching `trick_nextflow_cache.nf` [romantic_fourier] - revision: 22b984aca2 executor > slurm (2) [22/0f9755] process > make_a_large_file [100%] 1 of 1 ✔ [b1/d1cc88] process > inspect_large_file [100%] 1 of 1 ✔
Checking the work directories sizes reveals a ~1 Gb intermediate file generated by make_a_large_file():
[spvensko@c6145-2-9 tricking_nextflow_cache_example]$ du -hscL work/ 1.1G work/ 1.1G total
Now we run Nextflow script again and indicate that we'd like intermediate files to be deleted:
[spvensko@c6145-2-9 tricking_nextflow_cache_example]$ nextflow trick_nextflow_cache.nf -resume --delete_intermediates True N E X T F L O W ~ version 21.10.6 Launching `trick_nextflow_cache.nf` [exotic_jennings] - revision: 22b984aca2 executor > slurm (1) [22/0f9755] process > make_a_large_file [100%] 1 of 1, cached: 1 ✔ [b1/d1cc88] process > inspect_large_file [100%] 1 of 1, cached: 1 ✔ [16/f99612] process > clean_work_files (1) [100%] 1 of 1 ✔
Note the process clean_work_files() ran this time.
Checking the work directories again revealed a reduction from 1.1G to 60K! 🚀
[spvensko@c6145-2-9 tricking_nextflow_cache_example]$ du -hscL work/* 24K work/16 20K work/22 16K work/b1 60K total
This in itself is not too impressive - anyone can delete or replace a file in a work directory. The magical part happens when we re-run the workflow:
[spvensko@c6145-2-9 tricking_nextflow_cache_example]$ nextflow trick_nextflow_cache.nf -resume --delete_intermediates True N E X T F L O W ~ version 21.10.6 Launching `trick_nextflow_cache.nf` [mighty_albattani] - revision: 22b984aca2 [22/0f9755] process > make_a_large_file [100%] 1 of 1, cached: 1 ✔ [b1/d1cc88] process > inspect_large_file [100%] 1 of 1, cached: 1 ✔ [16/f99612] process > clean_work_files (1) [100%] 1 of 1, cached: 1 ✔
Nextflow pulled the make_a_large_file() process from cache despite the file emitted (make_a_large_file.out.a_large_file) being a 20K sparse file!
Next, let's go file-by-file and explain what's happening behind the scenes.
utilities.nf
#!/usr/bin/env nextflow
process clean_work_dirs {
input:
tuple val(directory)
output:
val(1), emit: IS_CLEAN
script:
"""
for dir in ${directory}; do
if [ -e \$dir ]; then
echo "Cleaning: \$dir"
files=`find \$dir -type f `
echo "Files to delete: \$files"
clean_work_files.sh "\$files" "null"
fi
done
"""
}
process clean_work_files {
cache 'lenient'
input:
val(file)
output:
val(1), emit: IS_CLEAN
script:
"""
clean_work_files.sh "${file}"
"""
}
utilities.nf consists of two process - clean_work_dirs and clean_work_files. clean_work_dirs appears to delete the contents of a work directory while clean_work_files targets specific files for deletion. Personally, I have only been using clean_work_files within my workflows. I have included these processes into their own module to allow easy aliasing when including the process into main.nf. For example, consider our current example in which we're running a single cleaning process. Defining the cleaning process within main.nf wouldn’t be the worst idea in the world. However, once you start cleaning multiple outputs files from multiple processes, having multiple cleaning processes defined in main.nf (as opposed to using aliasing while making include statements - see below) get messy fast.
include { clean_work_files as clean_trimmed_fastqs } from '../utilities/utilities.nf'
include { clean_work_files as clean_sorted_bams } from '../utilities/utilities.nf'
...
include { clean_work_files as clean_files_n} from '../utilities/utilities.nf'
clean_work_files.sh
#!/bin/bash
# https://raw.githubusercontent.com/SystemsGenetics/GEMmaker/master/bin/clean_work_files.sh
# This script is meant for cleaning any file in a Nextflow work directory.
# The $files_list variable is set within the Nextflow process and should
# contain the list of files that need cleaning. This can be done by creating
# a channel in a process that creates files, and merging that channel with
# a signal from another process indicating the files are ready for cleaning.
#
# The cleaning process empties the file, converts it to a sparse file so it
# has an acutal size of zero but appears as the original size, the access
# and modify times are kept the same.
files_list="$1"
for file in ${files_list}; do
# Remove cruff added by Nextflow
file=`echo $file | perl -p -e 's/[\\[,\\]]//g'`
if [ -e $file ]; then
# Log some info about the file for debugging purposes
echo "cleaning $file"
stat $file
# Get file info: size, access and modify times
size=`stat --printf="%s" $file`
atime=`stat --printf="%X" $file`
mtime=`stat --printf="%Y" $file`
# Make the file size 0 and set as a sparse file
> $file
truncate -s $size $file
# Reset the timestamps on the file
touch -a -d @$atime $file
touch -m -d @$mtime $file
fi
done
clean_work_files.sh is self-explanitory thanks to its great inline documentation.
trick_nextflow_cache.nf
nextflow.enable.dsl=2
include { clean_work_files } from './utilities.nf'
params.delete_intermediates = ''
process make_a_large_file {
cache 'lenient'
output:
tuple val("foo"), path("1G_file"), emit: a_large_file
script:
"""
dd if=/dev/zero of=1G_file bs=1G count=1
"""
}
process inspect_large_file {
cache 'lenient'
input:
tuple val(samp), path(required_input_file)
output:
tuple val(samp), path("file_stats"), emit: file_stats
script:
"""
ls -ldhrt ${required_input_file} > file_stats
"""
}
workflow {
make_a_large_file()
inspect_large_file(
make_a_large_file.out.a_large_file)
make_a_large_file.out.a_large_file
.join(inspect_large_file.out.file_stats, by: [0])
.flatten()
.filter{ it =~ /_file$/}
.set{ large_file_done_signal }
if( params.delete_intermediates ) {
clean_work_files(
large_file_done_signal)
}
}
trick_nextflow_cache.nf begins with a shebang, a standard inclusion statement (to import the cleaning process), and a parameter definition used for triggering intermediate file deletion. Next are two separate processes, the latter dependent on output from the former. The first process, make_a_large_file(), generates a one gigabyte sized file which is emitted through the a_large_file channel. The second process, inspect_large_file(), takes the emittied channel and simply ls -lhdrts it. In this example, you can imagine make_a_large_file() being an aligner process (e.g. bwa) and inspect_large_file() as being a variant caller (e.g. stelka2).
Once the inspect_large_file() process has completed, the actual intermediate large file itself is no longer needed. That is where the following code block comes into play:
make_a_large_file.out.a_large_file
.join(inspect_large_file.out.file_stats, by: [0])
.flatten()
.filter{ it =~ /_file$/}
.set{ large_file_done_signal }
...
if( params.delete_intermediates ) {
clean_work_files(
large_file_done_signal)
}
In this block, we are taking the channel containing the intermediate file we want to delete (the 1 Gb file), joining it to the channel containing the eventual endpoint file (file_stats). Note we are joining them on the 0th element which is the samp variable. We want to ensure that the intermediate file we're deleting and the corresponding output file are linked by a sample-level identifier. Once we have the joined tuple, we flatten() it and filter{} each element for the intermediate file suffix we are seeking to delete (_file$ in this case). We then pass this new tuple of deletable intermediates to clean_work_files() assuming params.delete_intermediates is true.
Note: Be as specific as possible with your filter{} regex string!!!
Limitations and Pitfalls
I encountered several hurdles when implementing this strategy into our workflows. I've described them below so others can avoid them. Please let me know of any other examples the community encounters and I'll include them.
Limits workflow expansion
This intermediate file deletion strategy, in its current form, should really only be applied to workflows that are well-established end-to-end. Deletion of intermediate files will result in prevention of workflow expansion. For example, consider a scenario in which you have alignment files and are performing variant calling with subsequent BAM cleanup:
process align {
...
}
process variant_caller_x {
...
}
workflow {
main:
align()
variant_caller_x(align.out.alns)
align.out.alns
.join(variant_caller_x.out.vcfs, by: [0])
.flatten()
.filter{ it =~ /.bam$/ }
.set{ cleanable_bams }
clean_work_files(
cleanable_bams)
}
After completing this workflow, you have your variant files and your storage hungry intermediate BAM files will be eliminated as expected.
Now, let's assume a colleague comes along and suggests also running the latest and greatest variant_caller_y (in addition to your original variant_caller_x). You include the variant_caller_y() process call to workflow (see below) and re-run the Nextflow:
process align {
...
}
process variant_caller_x {
...
}
process variant_caller_y {
...
}
workflow {
main:
align()
variant_caller_x(align.out.alns)
variant_caller_y(align.out.alns)
align.out.alns
.join(variant_caller_x.out.vcfs, by: [0])
.join(variant_caller_y.out.vcfs, by: [0])
.flatten()
.filter{ it =~ /.bam$/ }
.set{ cleanable_bams }
clean_work_files(
cleanable_bams)
}
However, upon running the above code you will be greeted with a curious error. What gives?! The deletion of the intermediate BAM file from the initial run means you can no longer use those BAMS to call variants. Nextflow is unaware that the BAM files are effectively useless since, from its perspective, they are perfectly fine cached files. At this point, you realize you must delete the work directory associated with align() and re-run that portion of the workflow.
Checking for every downstream output
Another consideration is that one must ensure all downstream processes have performed and completed their work prior to deletion of the intermediate file. In other words, the workflow developer must perform all process and channel accounting before allowing clean_work_files to execute. This will likely result in some rather beastly join statements that will clutter the code. Luckily, Ben Sherman (@bentsherman) is working on updating Nextflow to do this accounting internally and removing this burden from the owrkflow developer.
Nanoseconds count!
Astute readers may note my excessive use of the cache directive in process definitions. Inclusion of this directive is required to overcome the next pitfall I encountered. Specifically, it appears the Access and Modify time modications performed on the sparse file by clean_work_files.sh are only to the nearest second, but my cluster's filesystem seems to track Access and Modify times down to sub-second resolution. As a result, the sparse file generated did not pass Nextflow's caching sniff test and results in the process being re-ran (with sparse files as input). The cache directive (when paired with the 'lenient' option) bypasses this issue by removing the Access and Modify times from the Nextflow caching equation.
Original stat output prior to cleaning (from .command.log):
[spvensko@c6145-2-9 dd915decbfc0f0992acd0454f7e14f]$ cat .command.log cleaning /PATH/TO/work/71/8fb6ae67536c55ac80815da9c4a231/a_fastq_2.trimmed.fq.gz File: ‘/PATH/TO/work/71/8fb6ae67536c55ac80815da9c4a231/a_fastq_2.trimmed.fq.gz’ Size: 4891699623 Blocks: 9591736 IO Block: 65536 regular file Device: 31h/49d Inode: 9248463003887304838 Links: 1 Access: (0644/-rw-r--r--) Uid: (321499/spvensko) Gid: ( 3150/vincent_lab) Context: system_u:object_r:nfs_t:s0 Access: 2022-12-14 23:49:37.616634000 -0500 Modify: 2022-12-15 00:02:27.847675000 -0500 Change: 2022-12-15 00:02:27.847675000 -0500 Birth: -
stat output of sparse file which replaced the original file:
[spvensko@c6145-2-9 dd915decbfc0f0992acd0454f7e14f]$ stat /PATH/TO/work/71/8fb6ae67536c55ac80815da9c4a231/a_fastq_2.trimmed.fq.gz File: ‘/PATH/TO/work/71/8fb6ae67536c55ac80815da9c4a231/a_fastq_2.trimmed.fq.gz’ Size: 4891699623 Blocks: 0 IO Block: 65536 regular file Device: 31h/49d Inode: 9248463003887304838 Links: 1 Access: (0644/-rw-r--r--) Uid: (321499/spvensko) Gid: ( 3150/vincent_lab) Context: system_u:object_r:nfs_t:s0 Access: 2022-12-14 23:49:37.000000000 -0500 Modify: 2022-12-15 00:02:27.000000000 -0500 Change: 2022-12-15 01:21:02.845392000 -0500 Birth: -
Note that the Access time from the original file (2022-12-14 23:49:37.616634000 -0500 ) is not the same as the Access time from the sparse file created by clean_work_files.sh (2022-12-14 23:49:37.000000000 -0500).
Target the file, not the file's symlink
The final hurdle I had to overcome was one of misdirection. Nextflow relies heavily upon symlinks when operating on local cluster filesystems to prevent excessive copying of files among work directories. As a result, it's possible for a channel element containing what appears to be a file's path to instead be a symlink pointing to that original file. Consider the following example:
nextflow.enable.dsl=2
process make_a_large_file {
cache 'lenient'
output:
tuple val("foo"), path("1G_file"), emit: a_large_file
script:
"""
dd if=/dev/zero of=1G_file bs=1G count=1
"""
}
process inspect_large_file {
cache 'lenient'
input:
tuple val(samp), path(required_input_file)
output:
tuple val(samp), path(required_input_file), path("file_stats"), emit: file_stats
script:
"""
ls -ldhrt ${required_input_file} > file_stats
"""
}
workflow {
make_a_large_file()
make_a_large_file.out.a_large_file.view()
inspect_large_file(
make_a_large_file.out.a_large_file)
inspect_large_file.out.file_stats.view()
}
Running this script results in the following output:
[spvensko@c6145-2-9 tricking_nextflow_cache_example]$ nextflow real_vs_symlink.nf -resume N E X T F L O W ~ version 21.10.6 Launching `real_vs_symlink.nf` [pedantic_wright] - revision: fffa5484af [22/0f9755] process > make_a_large_file [100%] 1 of 1, cached: 1 ✔ [b1/d1cc88] process > inspect_large_file [100%] 1 of 1, cached: 1 ✔ [foo, /home/spvensko/tricking_nextflow_cache_example/work/22/0f975597019f5e81e7561400e3d7b0/1G_file] [foo, /home/spvensko/tricking_nextflow_cache_example/work/b1/d1cc88cf2ba4b16eb770d90b09f4aa/1G_file, /home/spvensko/tricking_nextflow_cache_example/work/b1/d1cc88cf2ba4b16eb770d90b09f4aa/file_stats]
Notice that both make_a_large_file()'s and inspect_large_files()'s emitted channels contain a path to 1G_file. However, upon closer inspection, we realize one path is the actual file while the other is simply a symlink to the original file:
[spvensko@c6145-2-9 tricking_nextflow_cache_example]$ file /home/spvensko/tricking_nextflow_cache_example/work/22/0f975597019f5e81e7561400e3d7b0/1G_file /home/spvensko/tricking_nextflow_cache_example/work/22/0f975597019f5e81e7561400e3d7b0/1G_file: data [spvensko@c6145-2-9 tricking_nextflow_cache_example]$ file /home/spvensko/tricking_nextflow_cache_example/work/b1/d1cc88cf2ba4b16eb770d90b09f4aa/1G_file /home/spvensko/tricking_nextflow_cache_example/work/b1/d1cc88cf2ba4b16eb770d90b09f4aa/1G_file: symbolic link to `/home/spvensko/tricking_nextflow_cache_example/work/22/0f975597019f5e81e7561400e3d7b0/1G_file'
In this case 1G_file was effectively passed through inspect_large_file(), but a symlink to the original file was emitted rather than the original file itself.
So, what does this matter?
If the workflow developer is not careful and deletes the symlink to 1G_file (../work/b1/d1cc88cf2ba4b16eb770d90b09f4aa/1G_file) rather than the 1G_file itself (../work/22/0f975597019f5e81e7561400e3d7b0/1G_file) then they will not save any space and instead disrupt the caching of their workflow.
The moral of this story is to ensure the channel and element you are targeting with clean_work_files is the actual file you want to delete and not a symlink to that file.
Conclusion
I hope this post helps in explaining why users may want to delete intermediate files in Nextflow, how to perform intermediate deletion, and some of the limitations and pitfalls associated with this approach.
Please do not hesitate to contact me with any thoughts or comments at steven_vensko@med.unc.edu or @spvensko on the nextflow.slack.com.