Open Source Computational Tools
PIRL uses and creates a variety of computational tools for our work in genomics and computational immunology. Some of these tools are carried over from previous work by the Vincent Lab at UNC, and OpenVax. Newer tools which are developed by PIRL will be listed below.
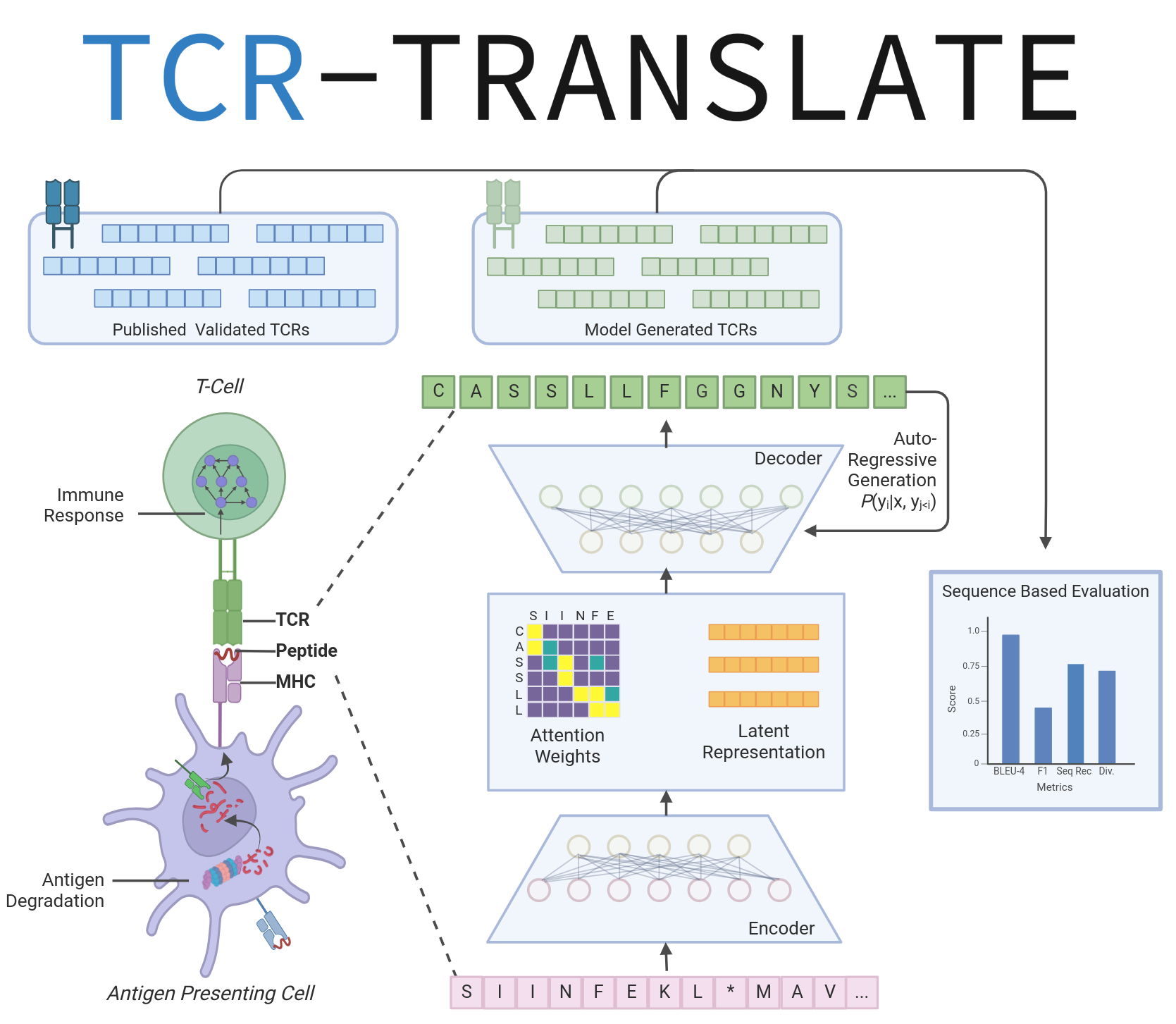
A sequence-to-sequence language model for antigen-specific TCR design that addresses data sparsity through techniques from low-resource machine translation.
Tumor antigen prediction pipeline used to curate tumor-specific predicted MHC ligands from a variety of mutational and expression sources.
Multiplex ELISpot assay design and deconvolution tool for determining epitope-specific T-cell responses from a large number of candidate peptides.
A tricky part of constructing large immunological datasets is dealing with the chaotic variability in naming of MHC molecules across species and within different research communities. MHCgnomes is a parser for MHC nomenclature which uses several reference data sources and a robust parsing strategy to standardize MHC nomenclature from a variety of different naming schemes you might encounter in the wild.